Abstract
A disposable card incorporating specific DNA probes targeting the 16 S rRNA gene of Streptococcus suis was developed for magnetically labeled target DNA detection. A single-stranded target DNA was hybridized with the DNA probe on the SPA/APTES/PDMS/Si as-prepared card, which was subsequently magnetically labeled with superparamagnetic beads for detection using an anisotropic magnetoresistive (AMR) sensor. An almost linear response between the output signal of the AMR sensor and amount of single-stranded target DNA varied from 4.5 to 18 pmol was identified. From the sensor output signal response towards the mass of magnetic beads which were directly immobilized on the disposable card surface, the limit of detection was estimated about 312 ng ferrites, which corresponds to 3.8 μemu. In comparison with DNA detection by conventional biosensor based on magnetic bead labeling, disposable cards are featured with higher efficiency and performances, ease of use and less running cost with respects to consumables for biosensor in biomedical analysis systems operating with immobilized bioreceptor.
Export citation and abstract BibTeX RIS

Original content from this work may be used under the terms of the Creative Commons Attribution 3.0 licence. Any further distribution of this work must maintain attribution to the author(s) and the title of the work, journal citation and DOI.
1. Introduction
Thanks to the immense advancement of materials science and engineering in the last decades, a great diversity of biosensors has been developed on the basis of electrochemical, magnetic, magneto-electric as well as optical principles. This exhibits great applications in all fields of the human life, from food safety, medical diagnostics, all kinds of process control in industrial, agricultural and other productions to environment control and monitoring, and even to bio-terrorism countering [1–3]. Although commercial sensors and devices have been widely available for efficient uses, a great deal of intensive research is in progress for further development. Beside approaches to higher sensitivity, higher resolution and reliability of the systems are desired, the concerns of development currently focus primarily on higher efficiency and performances, robustness, compactness, the ease of uses, environment and running cost friendly especially with respects to power efficiency, reagents and other consumable consumption.
The use of the biosensors in the medical engineering and health care is integrated in so-called medical analysis systems. In such a system, a biosensor usually is structured with three parts. The first one is a selective biological element usually called a bioreceptor which consists of micro-organisms, enzymes, antibodies or nucleic acids etc. The second is a transducer which is of electrochemical, optical, magnetic, piezoelectric nature and has the function to transform the signal resulted from the interaction between the analytes and the biological elements into a signal that can be measured and quantified. The third part is a signal processor that provides a user-friendly signal display. In conventional biosensors, the biological element is usually directly immobilized on the sensor surface. Reusing the biosensor always involve a complex procedure of cleaning the analytes of the previous measurement from the biosensor surface which is considerably difficult to achieve completely due to the strong and specific interaction between the analytes and the bioreceptors. Due to these obstacles, there is a growing tendency to use disposable cards incorporated to the biosensor in biomedical analysis systems operating with immobilized bioreceptor [4, 5].
In spintronics based biochip applications, the magnetic microbeads or nanoparticles are usually labeled with biomolecules (proteins, DNA etc) and are employed in the detection of target biomolecules by binding the probe biomolecules immobilized on the magnetic sensing surface. In in-vivo applications, moreover, magnetic nanoparticles are usually located in deep tissue. Thus, distant magnetic nanoparticle detections are required. In general, the non-invasive bio-detection has attracted a lot of attention over the years [6, 7]. Among them, the DNA-magnetic bead detection using disposable cards is a suitable approach.
Recently, our group has successfully detected magnetic nanoparticles which directly immobilized on the simple anisotropic magnetoresistive (AMR) sensor surface [8]. This work develops and demonstrates the proof-of-concept of the AMR based biosensor operating with disposable cards incorporating specific DNA probes targeting the 16 s rRNA gene of Streptococcus suis for the detection of magnetically labeled target DNA.
2. Experimental
2.1. Fabrication of disposable card
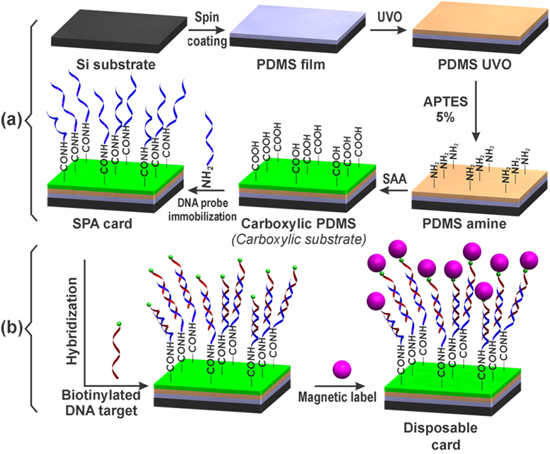
The disposable card (SPA) was fabricated following the experimental process as shown in figure 1(a). Here, a polydimethylsiloxane (PDMS) thin layer was first deposited by spin coating on silicon substrate, followed by ultraviolet/ozone (UVO) treatment, 3-aminopropyltriethoxy (APTES) grafting, functionalizing using succinic acid anhydride (SAA) and SPA probe immobilizing.
Figure 1. (a) The fabrication process of the SPA card and (b) DNA target hybridization and magnetic label on the SPA card.
Download figure:
Standard image High-resolution imageAt first, the PDMS monomer (Dow Corning Sylgard-184 PDMS) was mixed in a glass container with a curing agent with a weight ratio of 10:1 and pumped vacuum for 15 min to remove air bubbles. It was subsequently spread on the silicon substrate by spin coating (SUSS MicroTec spin coaters) at a speed of 6000 rpm for 10 min and then cured at 70 °C for one hour. After washed with pure ethanol and dried with pure nitrogen gas, the PDMS film was treated by UVO in UVO chamber (Jelight Corp. model 144AX-220) for 20 min (called PDMS UVO). This PDMS UVO was immersed in APTES: ethanolsolution (5:100 v/v) for 15 min. After rinsing in ethanol and a subsequent heated at 100 °C for one hour, the surface of APTES treated PDMS (also called PDMS amine) was modified by dropping the 200 mM SAA (pH 6) solution on its. The SAA coated PDMS amine was spotted with a solution of the 2 μM DNA probe (SPA, Intergrated DNA Technology) and 1 mg ml−1 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) in 100 mM 2-(N-morpholino) ethanesulfonic acid (MES) buffer pH 5.5 and was subsequently incubated at 25 °C for two hours. After two times being washed with 250 mM tris pH 8 and 0.01% tween-20 (TT) buffer and then with deionized (DI) water, the SPA immobilized card is now called SPA card. We note here that after each functionalization step as described above, a Fourier transform infrared spectroscopy (FTIR) spectrum was taken on a Shimadzu IR Prestige-21 system.
2.2. Preparation of DNA hybridization assay and magnetic label on the SPA card
Different amount of biotinylated single-stranded target DNA (DNA(SS)) of 4.5, 9 and 18 pmol in hybridization buffer of 300 mM NaCl, 30 mM citrate sodium, 0.1% v/v sodium dodecyl sulfate (SDS), pH 7 was spotted onto SPA card and incubated in Hybridizer HB-1000 (UVP) at 50 °C for one hour for DNA hybridization (see figure 1(b)). The SPA card was washed two times with washing buffer SSC (150 mM NaCl, 15 mM citrate sodium, 0.1% SDS, pH 7) and DI water. The streptavidin conjugated superparamagnetic beads (Invitrogen) (1 μm in diameter) contains a monolayer of streptavidin covalently coupled to the surface of the beads. This leaves the vast majority of biotin binding sites for the binding of the biotinylated target DNA(SS). 3 μg of magnetic beads were incubated on the SPA card at 25 °C for 15 min with the hybridized target DNA(SS) in binding and washing buffer (5 mM tris-HCl pH 7.5, 500 μM EDTA, 1 M NaCl) (see in figure 1(b)). Unbound magnetic beads were removed from the SPA card by washing three times with binding and washing buffer. SPA card with magnetically labeled target DNA was subsequently inserted into the AMR sensor for signal detections.
2.3. Sensor fabrication
The AMR sensor was fabricated following the procedure and technical features described in [8], where Wheatstone bridges incorporating a serially connected ensemble of simple AMR elements of 5 nm thick Ni80Fe20 film were produced by using the magnetron sputtering equipment (model ATC 2000) and the UV lithographer (model MJB4) at the VNU Key Laboratory for Micro-Nanotechnology. This Ni80Fe20 film was grown on SiO2/Si for 5 min with the deposition rate of 1 nm min−1. In this case, the magnetic anisotropy was established thanks to the application of a pinned magnetic field along the sensing magnetoresistor length. For an acceptable sensitivity, here, the magnetoresistive element with length l = 4 mm and width w = 150 μm was used.
2.4. Characterization techniques
The characterization of the card substrate after each functionalization step and SPA probe immobilization was performed by means of IR Prestige-21 Shimadzu Fourier transform infrared spectrophotometer at a grazing angle of 80° with the resolution 4 cm−1. The water contact angle (WCA) was measured using a simple experimental apparatus described by Lamour et al [9] with a digital camera and the ImageJ software. The contact angles were recorded by dispensing 20 μl of DI water on the substrates with an error of ca. 2.5°. Surface morphology of the PDMS film and the SPA card were characterized by a Hitachi S4800 field-emission scanning electron microscope.
For the magnetoresistance measurement, the dc precision current source was supplied by using Keithley 6220 and the output voltage (Vout) was recorded by Keithley 2000 multimeter. The Vout voltage of the Wheatston bridge was detected by a DSP lock-in amplifier (model 7265 of Signal Recovery) in combination with an oscilloscope (Tektronic DP 4032) [8].
3. Results and discussion
3.1. Characterizations of substrates and the SPA card
The FTIR spectra taken on the card substrate after each functionalization step are presented in the figure 2. The FTIR spectrum of the PDMS film exhibits three peaks at 806 cm−1, 1088 cm−1 and 1258 cm−1 corresponding to the attribution of the Si-CH3, Si-O-Si and Si-CH3 stretching deformation vibration, respectively [10]. The peak with wavelength of 1088 cm−1 disappeared in the FTIR spectrum of the PDMS UVO. In addition, a new peak appears at 3669 cm−1. These findings are ascribed being connected to the destruction of Si-CH3 bonds and the formation of Si-OH [11, 12].
Figure 2. FTIR spectra of PDMS film before and after each functionalization step.
Download figure:
Standard image High-resolution imageFor the PDMS amine, the FTIR spectrum is featured with a rather weak peak at 1539 cm−1, which can be assigned to the N-H stretching vibration in the amino group of the grafted APTES. Finally, the FTIR spectrum of the carboxylic PDMS shows two more new peaks at 1643 cm−1 and 3449 cm−1, which are due to the amide vibration and the O-H vibration in the carboxylic groups, respectively. This indicates that the SSA has reacted with the amine on the PDMS amine to form amide and free carboxylic groups. These results confirm that the card substrate was successfully functionalized.
WCA of the card substrates were measured after each functionalization step. The WCA were estimated as 40°, 110°, 72° and 83° for silicon substrate, PDMS, PDMS UVO and carboxylic PDMS cards, respectively (see figure 3). The observed significant change of WCA as observed reveals the modification of the substrate surfaces. These results are comparable with those reported in [13, 14].
Figure 3. Values of the water contact angles at different fabrication steps of the card substrate.
Download figure:
Standard image High-resolution image3.2. DNA probe design and immobilization
The DNA probe (called SPA) specific for the 16 S rRNA gene of Streptococcus suis was designed on the basis of the Genbank of National Center for Biotechnology Information (NCBI) and by means of the Clustal X 2.0 program for DNA sequence alignment. The obtained SPA sequence is illustrated in figure 4. The SPA has an amino group at the 5' end for the probe immobilization on the carboxylic substrate. In addition, the SPA also contains the 20 thymidines (PolyT) at the 5' end for reducing the steric hindrance of the card surface to DNA hybridization. In addition, a model synthetic single-stranded target DNA(SS) with biotin at the 5' end and a control DNA(SF) without biotin are also given in figure 4 below. The biotin at the 5' end of the single-stranded was used for binding to the streptavidin conjugated magnetic beads. The streptavidin-biotin complex is the strongest known non-covalent interaction between a protein and a ligand. The bond between biotin and streptavidin is formed rapidly and robustly. These features of biotin and streptavidin are useful for conjugation of biotinylated DNA and streptavidin conjugated magnetic beads.
Figure 4. DNA sequences of the DNA probe, model target DNA and control DNA.
Download figure:
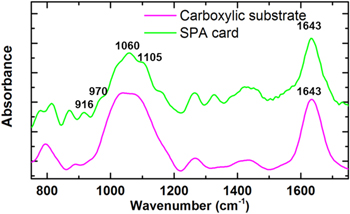
Standard image High-resolution imageThe DNA probe SPA was immobilized on the carboxylic substrate (figure 1(a)) by a chemical method using the water soluble carboxyl-to-amino crosslinker EDC. The low wavenumber FTIR spectra of the carboxylic substrate and the SPA card are shown in figure 5. In comparison with the spectrum of the carboxylic substrate (see also figure 2), the additional four new peaks appeared at 916 cm−1, 970 cm−1, 1060 cm−1 and 1105 cm−1 in the SPA card spectrum, which correspond to the DNA ribose-phosphate skeletal motions, ribose C−O and P−O−C stretching, respectively [15]. These findings suggest that the target DNA is well immobilized on the carboxylic substrate.
Figure 5. Low wavenumber FTIR spectra of card substrate and the SPA card.
Download figure:
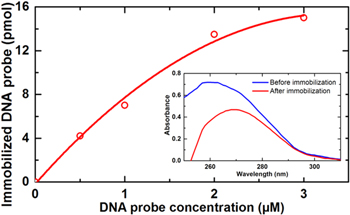
Standard image High-resolution imageThe DNA probe concentration was determined by Thermo Scientific NanoDrop 2000 spectrophotometer using DNA absorbance at 260 nm. Shown in the inset is the UV–vis absorption spectra taken on the DNA probe before (the blue curve) and after (the red curve) immobilizing on the card substrate. It is seen that, the latter absorption peak at 260 nm of DNA probe is clearly reduced with respect to the former one, indicating the immobilization of the DNA probe on the card substrate. The DNA probe concentration dependence of the immobilized DNA probe is presented in figure 6. There, the optimization of the immobilized DNA probe can be determined. Indeed, it can be seen from this figure that a tendency to saturation is reached at the DNA probe concentration higher than 2 μM. Thus, one can estimate that the appropriate DNA probe concentration for immobilizing on the card substrate is 2 μM. At this concentration immobilized DNA probe amount on the card substrate reached the value of 13.5 pmol.
Figure 6. Optimization graph of the DNA probe concentration for immobilization on the card substrate. Inset: absorption spectra of the DNA probe SPA before and after immobilization on the card substrate.
Download figure:
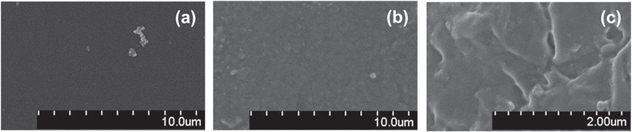
Standard image High-resolution imageFigure 7 presents surface morphology of the PDMS film and the SPA card. Compared to the initial smooth surface of the PDMS film (figure 7(a)), the SPA card has rough micro textured surface (figures 7(b) and (c)) due to UVO treatment. This is in good agreement with the report [16].
Figure 7. SEM images of surfaces of (a) the PDMS film, and (b), (c) the SPA card.
Download figure:
Standard image High-resolution image3.3. Detection system setup and signal measurement
In this investigation, the DNA was magnetically labeled by using superparamagnetic 1 μm Dynabeads®MyOne™ streptavidin C1 (with 26% ferrites). Room temperature magnetic hysteresis loop of the SPA card with an amount of 30 μg magnetic beads measured on the Lake Shore 7404 vibration sample magnetometer (VSM) is shown in figure 8. It can be seen that the saturation magnetization reaches the value as high as 23 emu g−1. At a low magnetic field of 30 Oe, the magnetization is only 2.86 emu g−1. The superparamagnetic behavior of the magnetic beads was confirmed by the negligible coercivity and remanence (see the inset of figure 8).
Figure 8. Magnetization curve of a disposable card involving DNA probes with an amount of 30 μg of superparamagnetic 1 μm Dynabeads® MyOne™ streptavidin C1.
Download figure:
Standard image High-resolution imageThe hybrid DNA-magnetic beads were detected by means of the simple AMR sensors in a Wheatstone bridge as described in section 2.3. The sensor characteristics are illustrated in figure 9. Clearly, the sensor is rather sensitive below the applied magnetic field of 10 Oe. Indeed, the maximum sensitivity of 3.6 mV Oe−1 can be achieved at an applied bias magnetic field as low as H = −5.4 Oe. This is considered as a good working point of the bias magnetic field for magnetic beads detection in experiments.
Figure 9. Magnetic field dependence of (a) output voltage and (b) relative derivative of AMR sensor.
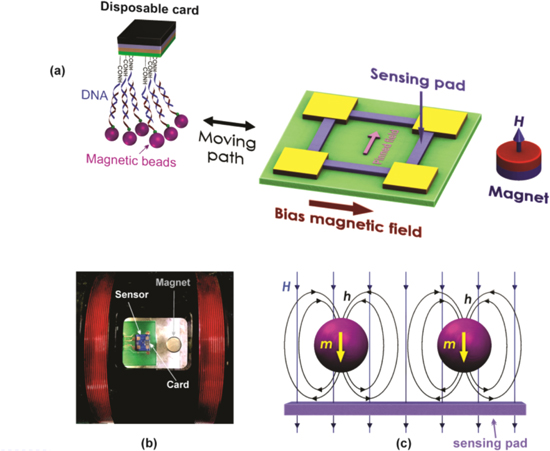
Download figure:
Standard image High-resolution imageThe measurement setup for DNA-magnetic bead detection is shown in figures 10(a) and (b). The SPA card with the magnetic beads was placed at a distance of 10 μm from the sensor surface. In this configuration, the magnetic bead is vertically magnetized thanks to the magnetic field (H) of about 30 Oe generated by the permanent magnet. Its stray fields (h), however, are mainly distributed in the plane of the active sensing magnetoresistor (figure 10(c)). Thus, the magnetic beads are magnetically enabled enable to be detected by the sensor.
Figure 10. (a) The scheme and (b) image of the experimental setup for the disposable card detection using the AMR sensor and magnetic fluxes created by permanent magnet H and (c) magnetic particle h.
Download figure:
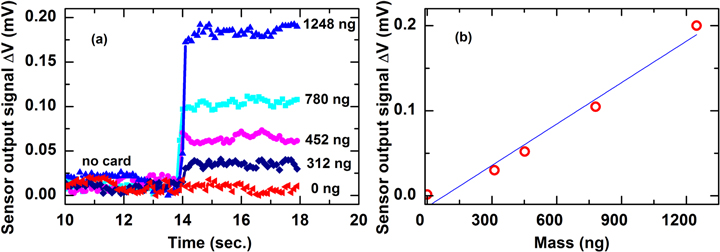
Standard image High-resolution imageThe detection of the magnetic beads was performed by recording the sensor signal for the case of with and without disposable cards. The obtained results for different disposable cards with different amounts of magnetic particles varied from 0 to 1248 ng are presented in figure 11(a). The smallest detectable amount of magnetic particles was estimated about 312 ng which corresponds to a detection limit of 3.8 μemu. The mass dependence of the output voltage signal exhibits an almost linear behavior with a slope of 181 μV μg−1 (see figure 11(b)).
Figure 11. The sensor signals recorded disposable cards with different amount of (a) magnetic particles and (b) magnetic particles mass-dependence signal strength.
Download figure:
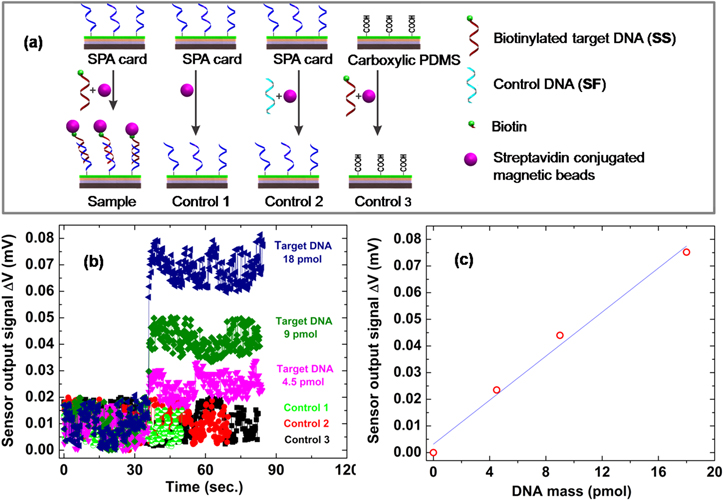
Standard image High-resolution imageThe SPA cards with different amount of magnetically labeled single-stranded target DNA (sample 1, 2 and 3) of 4.5, 9 and 18 pmol, respectively, were measured. For further verifying the measurement results, the same experiments were performed in different cards (see figure 12(a)) consisting of a SPA card without target DNA (control 1), a SPA card with control non-biotinylated DNA(SF) (control 2) and the card substrate without the SPA probe incubated with target DNA and magnetically labeled (control 3). The results in figure 12(b) clearly show that only the SPA card incubated with target DNA and subsequently magnetically labeled (samples) exhibits significant changes in the sensor signal. This signal changes seem to increase almost linearly with the increasing amount of target DNA (figure 12(c)). From these considerations, the detection limit is estimated of about 4.5 pmol single-stranded target DNA.
Figure 12. (a) A schematic representation of the target DNA bioassay and controls, (b) the sensor signals recorded for disposable cards with different amounts of single-stranded target DNA on the SPA card and controls and (c) target DNA mass-dependence of the sensor signal (the line is a guide to the eye).
Download figure:
Standard image High-resolution imageBased on this disposable card with SPA probe approach, the 16 S rRNA gene of Streptococcus suis could be detected. In addition, resolution of the biosensor system could be enhanced by developing a bioassay, combining target DNA recognition with rolling circle amplification before magnetic label [17] or choosing magnetic beads with proper sizes, higher magnetic moment and surface based binding ability [18, 19] in combination with enhancing the sensor sensitivity.
4. Conclusion
For a distant DNA-magnetic bead detection approach, the disposable card with DNA probes specific for the 16 S rRNA gene of Streptococcus suis was successfully designed, fabricated and investigated. Moreover, the proof-of-concept biosensor using an AMR sensor is demonstrated. The system is able to detect hybrid DNA-magnetic beads with the detection limit of 4.5 pmol of single-stranded target DNA. This novel approach opens up new possibilities for the development of highly sensitive, low-cost and rapid-detection DNA biosensor system.
Acknowledgments
This work was supported by Vietnam National University, Hanoi (VNU), under project No. QGTD 13.24 and QG.16.26.